
Teaching Summary (offerings vary year to year):
Undergraduate Research Projects (BIOL/BIOC 4900 Special Problems)
Plant Biochemistry and Biotechnology (BIOC 6650) graduate level only
Plant Physiology (BIOL 4503/5503) suitable for juniors, seniors and grad students
Genetics (BIOL 3451)
Biology of Higher Plants (BIOL 2241)
(If you are looking for course material, go to UNT CANVAS (unt.instructure.com)
Graduate and undergraduate research opportunities available: click here and inquire
Research Summary:
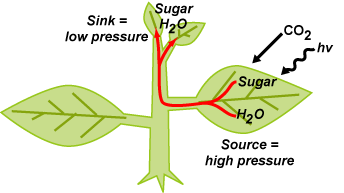
Research in my laboratory revolves around the phloem transport system of plants and
how it functions as a whole-plant communication network to enable disparate organs
to function as an integrated complete organism. Within this broad context, we have
projects in two main areas: 1) The role of the phloem in coordinating carbon metabolism
and nutrient utilization between photosynthetic source leaves and heterotrophic sink
organs and 2) the role of the phloem in transporting signaling molecules from leaves
to growing tissues to mediate source control of sink growth and development. Together,
these trajectories contribute to our understanding of how plants control biomass partitioning
and growth patterns on a whole-plant level. We approach our questions with modern
tools in biochemistry, molecular biology and plant biotechnology, but always retain
sight of the whole organism in our answers.
Our laboratory is part of UNT's BioDiscovery Institute which aims to delivers research solutions to underpin the utilization of biosystems for sustainable production of food/feed, polymers, construction materials, bioactive molecules, and biofuels.
Professional Experience:
2017-Present: Professor, BioDiscovery Institute and Dept. of Biological Sciences, University of North Texas
2019 (6 mo): Visiting Professor, Dept. of Genetics, University of Georgia, Athens GA; Host, Dr. Chung-Jui (CJ) Tsai
2010-2017: Associate Professor, Dept. of Biological Sciences, University of North Texas
2012 (7 mo): Visiting Scientist, Max Planck Institute of Molecular Plant Physiology, Dept of Metabolic Networks (AG Stitt), Potsdam-Golm, Germany
2003-2010: Assistant Professor, Dept. of Biological Sciences, University of North Texas
1998-2003: Research Associate, Dept of Plant Biology, Cornell University, Ithaca, NY
1995-1998: Postdoctoral Fellow, Medical Research Council Laboratory of Molecular Biology, Cambridge, England
1989-1995: PhD, Plant Mol Biol & Biotech, University of Alberta, Canada
1985-1989: BSc, Genetics, University of Manitoba, Canada
Current and Recent Research Funding:
-
McGarry RC, Shah J, Ayre BG. Generating pathogen- / pest-resistant non-GMO cotton through targeted genome editing of oxylipin signaling pathways. United States Department of Agriculture; National Institute for Food and Agriculture; Agriculture and Food Research Initiative Competitive Grants Program. AWARDED $294,000, Jan 2021 - Jan 2024.
- Ayre BG, McGarry RC, Eshed-Williams L (Hebrew University), van der Knaap E (University of Georgia). Elucidating and manipulating the CLAVATA-WUSCHEL circuit in cotton to understand meristem homeostasis in relation to fruit size and shape. United States-Israel Binational Agricultural Research and Development (BARD) Fund. AWARDED $310,000 Sept 2019 - July 2023.
- Ayre BG, McGarry RC. Redesigning the cotton plant's architecture to improve yield and quality. Cotton Incorporated: National Program. AWARDED ~$550,000, Jan 2010 - Jan 2023 (Renewed annually at ~$50,000).
- Ayre BG. Unraveling the link between carbohydrate transport and phosphate use: Can we improve carbon partitioning and reduce nutrient use? National Science Foundation, Directorate for Biological Sciences, Integrative Organismal Systems. AWARDED $564,315, Mar 2016 - Mar 2020.
Representative Manuscripts (Full list at Google Scholar):
- Yadav UP, Evers JF, Shaikh MA, Ayre BG (2022) Cotton phloem loads from the apoplast using a single member of its nine-member sucrose transporter gene family. J Exp Bot 73: 848-859. DOI: 10.1093/jxb/erab461.
- McGarry RC, Ayre BG (2021) Cotton architecture: examining the roles of SINGLE FLOWER TRUSS and SELF-PRUNING in regulating growth habits of a woody perennial crop. Curr Opin Plant Biol 59: 101968. DOI: 10.1016/j.pbi.2020.10.001.
- McGarry RC, Rao X, Li Q, van der Knaap E, Ayre BG (2020) SINGLE FLOWER TRUSS and SELF-PRUNING signal developmental and metabolic networks to guide cotton architectures. J Exp Bot 71: 5911-5923. DOI: 10.1093/jxb/eraa338
- Yadav UP, Shaikh MA, Evers J, Regmi KC, Gaxiola RA, Ayre BG (2019) Assessing long-distance carbon partitioning from photosynthetic source leaves to heterotrophic sink organs with photoassimilated [(14)C]CO2. In Methods Mol Biol, Vol 2014, pp 223-233. DOI: 10.1007/978-1-4939-9562-2_19
- Prewitt SF, Ayre BG, McGarry RC (2018) Cotton CENTRORADIALIS/TERMINAL FLOWER 1/SELF-PRUNING genes functionally diverged to differentially impact plant architecture. J Exp Bot 69: 5403-5417. 10.1093/jxb/ery324
- Ayre BG, Turgeon R (2018) Export of Photosynthates from the Leaf. In WWI Adams, I Terashima, eds, The Leaf: A Platform for Performing Photosynthesis. Springer International Publishing AG, pp 55-79. DOI: 10.1007/978-3-319-93594-2_3
- McGarry RC, Klocko AL, Pang M, Strauss SH, Ayre BG (2017) Virus-Induced Flowering: An Application of Reproductive Biology to Benefit Plant Research and Breeding. Plant Physiology 173: 47-55. DOI: 10.1104/pp.16.01336
- McGarry RC, Prewitt SF, Culpepper S, Eshed Y, Lifschitz E, Ayre BG (2016) Monopodial and sympodial branching architecture in cotton is differentially regulated by the Gossypium hirsutum SINGLE FLOWER TRUSS and SELF-PRUNING orthologs. New Phytologist 212: 244-258. DOI: 10.1111/nph.14037